I am a Computational Structural Biologist with strong expertise and deep interest in protein design, small molecule design, interactions and dynamics to engineer novel therapeutics. I am particularly passionate about projects that bridge the gap between computational predictions and experimental validation, finding true excitement in seeing computational designs come to life in the lab. I thrive on tackling complex biological challenges and use state-of-the-art computational approaches to engineer novel molecules that could make a meaningful difference in people's lives.
Previously, I worked as a Bioinformatician in the Bill and Melinda Gates Foundation-funded project at the National Institute of Health (NIH), applying computational modeling to study SARS-CoV-2 during the pandemic. My key contributions involved identifying viral variants of concern, modeling the destabilizing mutations in the Main protease (Mpro), and designing novel peptide antigens using cutting-edge de novo protein design techniques. Particulalry, this work further expanded my expertise in:
My journey, in the realm of bio-molecular structures started during my MS/MPhil studies at the National Center for Bioinformatics (NCB), where I worked on “Proteome-wide analysis for potential druggable candidates against Morganella morganii”, in which I completed my thesis in the Azam Lab. Before joining the Azam Lab, I was working as a Research assistant in Dasti Lab, where I was involved in bacterial cell culturing and sequence analysis of “CTXM-15 virulence profile of Uropathogenic E.coli (UPEC) strains” from Pakistan, which I submitted to Genbank NCBI.
I envision myself becoming a protein designer over the next decade, driven by the conviction that designing novel protein-architectures are the key to developing novel therapies for numerous diseases. My deep curiosity about these really fascinating structures, their interactions and dynamics fuels my passion for this cutting-edge field. The next ten years will be an exciting voyage for me. Now actively seeking roles in protein design
Discipline: Computational Structural Biology
Published the, PDB ID: 8WSV - Crystal structure HosA E.coli
Research Skills/Expertise
Laboratory: DNA/Plasmid extraction | PCR | Gel electrophoresis | Bacterial cell culturing | Broth & media preparation | Autoclaving | Centrifugation | Spectrophotometry | Microscopy
Computational:
Skilled in De novo protein/peptide design using techniques of BindCraft, RFDiffusion, Proteus, ProteinMPNN, DiffAb, Chroma, Boltz-1, Chai-1 etc
Proficient in molecular dynamics simulations(MDS) to study biomolecular structure, dynamics and interactions
Proficient in structural modeling of proteins, nucleic acids, membranes and their complexes
Experienced in mmgbsa/pbsa, ΔΔG, metadynamics and FEP+ analysis
Skilled in crystal structure refinement and analysis
Programming skills in Linux, Bash, R, NumPy, SciPy, Biopython, PyTorch, Docker, Anaconda, Jupyter Notebook and CUDA configuration --- Please for more refer to my CV
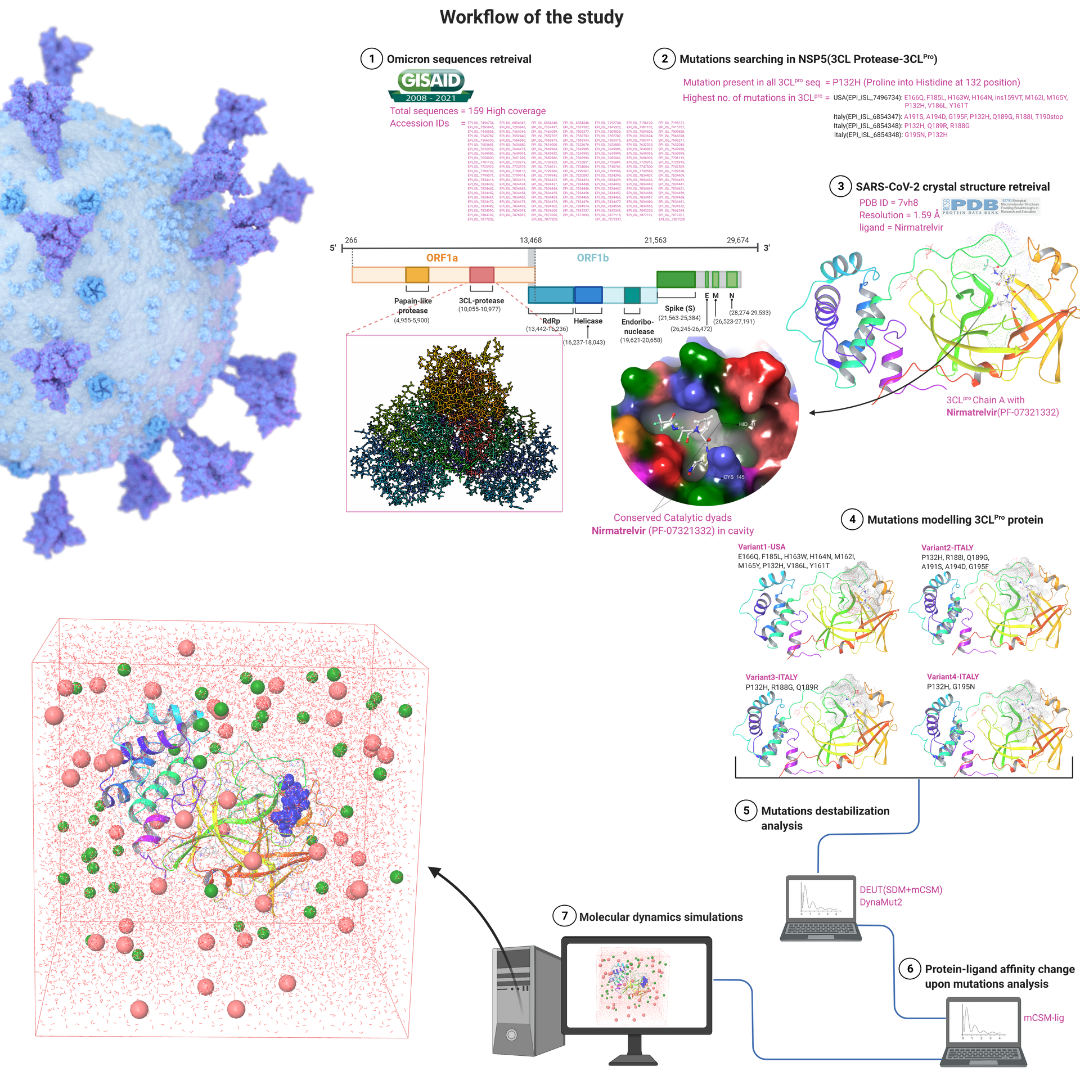
Samee’s most recent paper, Computational insights on the destabilizing mutations in the binding site of 3CL-protease SARS-CoV-2 Omicron (VOC) is now availabe at biorxiv preprint server.
Recent Posts
Check out the latest Papers, News, Events, and More »